miércoles, 22 de abril de 2020
Elevated rates of positive selection drive the evolution of pestiferousness in the Colorado potato beetle (Leptinotarsa decemlineata, Say)
Cohen et al., 2019
Insect pests are characterized by expansion, preference and performance
on agricultural crops, high fecundity and rapid adaptation to control
methods, which we collectively refer to as pestiferousness. Which
organismal traits and evolutionary processes facilitate certain taxa
becoming pests remains an outstanding question for evolutionary
biologists. In order to understand these features, we set out to test
the relative importance of genomic properties that underlie the rapid
evolution of pestiferousness in the emerging pest model: the Colorado
potato beetle (CPB), Leptinotarsa decemlineata Say. Within the Leptinotarsa genus, only CPB has risen to pest status on cultivated Solanum. Using whole genomes from ten closely related Leptinotarsa
species, we reconstructed a high-quality species tree of this genus.
Within this phylogenetic framework, we tested the relative importance of
four drivers of rapid adaptation: standing genetic variation, gene
family expansion and contraction, transposable element variation, and
protein evolution. Throughout approximately 20 million years of
divergence, Leptinotarsa show little evidence of gene family
turnover or transposable element variation contributing to pest
evolution. However, there is a clear pattern of pest lineages
experiencing greater rates of positive selection on protein coding
genes, as well as retaining higher levels of standing genetic variation.
We also identify a suite of positively selected genes unique to the
Colorado potato beetle that are directly associated with
pestiferousness. These genes are involved in xenobiotic detoxification,
chemosensation, and hormones linked with pest behavior and physiology.
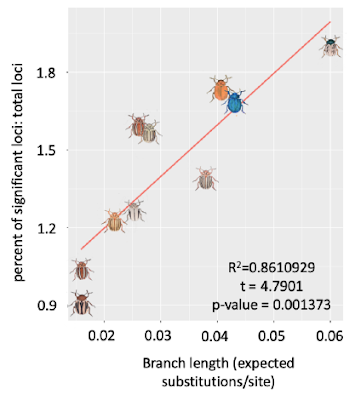
Proportion of genes under positive selection (results from adaptive branch-site random 880 effect model) to total genes explained by branch length with significant phylogenetic dependence, 881 Blomberg’s K (1.4593) > 1; PIC.variance p-value = 0.008991009.
.
Suscribirse a:
Enviar comentarios (Atom)










No hay comentarios:
Publicar un comentario