domingo, 3 de marzo de 2019
Large-scale replicated field study of maize rhizosphere identifies heritable microbes
William A. Walters et al. 2019.
Significance
In this very large-scale longitudinal field study of the maize rhizosphere microbiome, we identify heritable taxa. These taxa display variance in their relative abundances that can be partially explained by genetic differences between the maize lines, above and beyond the strong influences of field, plant age, and weather on the diversity of the rhizosphere microbiome. If these heritable taxa are associated with beneficial traits, they may serve as phenotypes in future breeding endeavors
Abstract
Soil microbes that colonize plant roots and are responsive to differences in plant genotype remain to be ascertained for agronomically important crops. From a very large-scale longitudinal field study of 27 maize inbred lines planted in three fields, with partial replication 5 y later, we identify root-associated microbiota exhibiting reproducible associations with plant genotype. Analysis of 4,866 samples identified 143 operational taxonomic units (OTUs) whose variation in relative abundances across the samples was significantly regulated by plant genotype, and included five of seven core OTUs present in all samples. Plant genetic effects were significant amid the large effects of plant age on the rhizosphere microbiome, regardless of the specific community of each field, and despite microbiome responses to climate events. Seasonal patterns showed that the plant root microbiome is locally seeded, changes with plant growth, and responds to weather events. However, against this background of variation, specific taxa responded to differences in host genotype. If shown to have beneficial functions, microbes may be considered candidate traits for selective breeding.
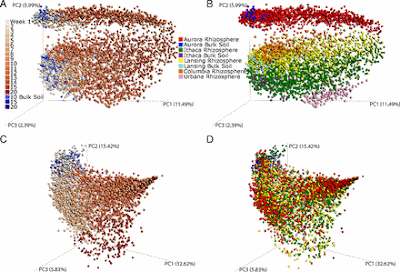
Environment and age strongly structure the rhizosphere microbial community. PCoA of unweighted (A and B) and weighted (C and D) UniFrac distances for maize rhizosphere and bulk soil samples. A and B show the same projection of the data, as do C and D. Symbols represent microbiomes and are colored by plant age (A and C; sampling week; see color gradient) by environment (B and D; by the specific field of origin). The first three PCs are plotted with the percentage of variation explained by each PC.
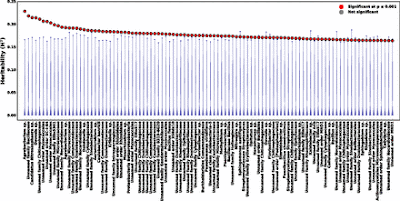
Broad-sense heritability of individual OTUs for the 2010 field study. The broad-sense heritability (H2) is shown for the 100 OTUs with highest H2 values. Circles show the actual H2 values for each OTU in decreasing order and blue distributions show the corresponding H2 values from 5,000 permutations of the data. Red circles indicate OTUs with P values ≤0.001. Taxonomies shown are most specific for each OTU.
Phylogeny of heritable OTUs. The tree shows the phylogenetic
relationships of OTUs found in 80% of all rhizosphere samples. Heritable
OTUs have branches colored in red. Major phyla are color-coded in the
outer circle, and the deepest named taxonomy is given for each OTU. The
tree is a subset of the Aug 2013 Greengenes tree. (Scale bar indicates
inferred mutation rate per site.).
.
.
Suscribirse a:
Enviar comentarios (Atom)











No hay comentarios:
Publicar un comentario